Access more sample types
Compatible with both FFPE and fresh frozen tissue samples.
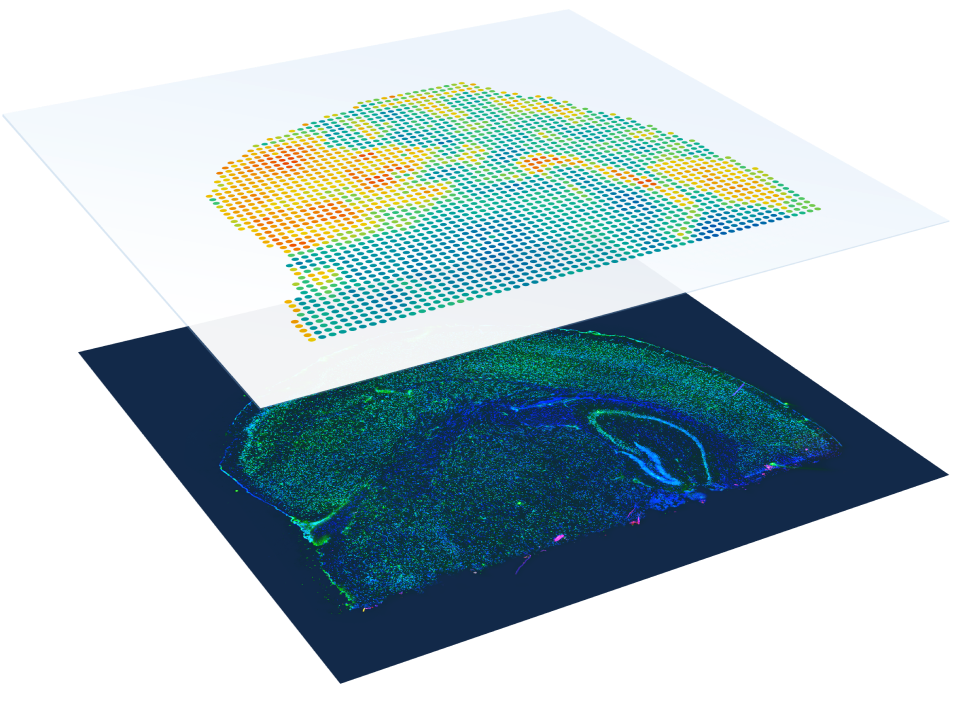
Whole tissue section profiling
No need to select regions of interest—analyze the whole transcriptome from an entire section.
High cellular resolution
1–10 cell resolution on average per spot depending on tissue type.
Diverse sample compatibility
Demonstrated data on a diverse set of organs across species (human, mouse, rat, and more).
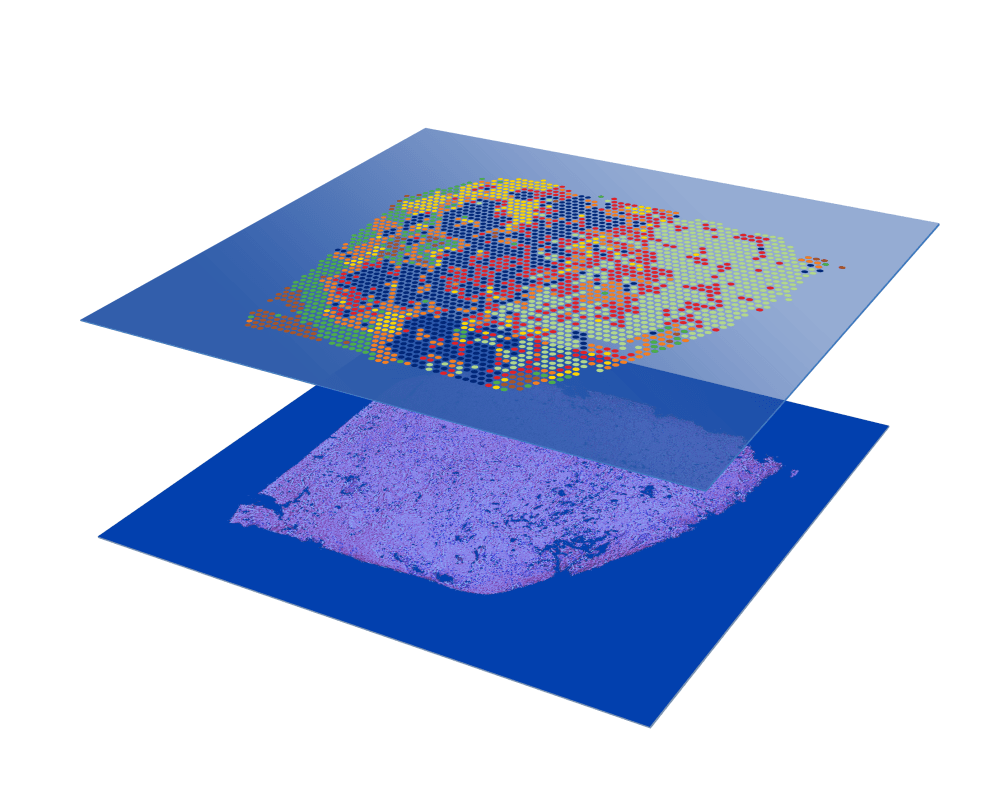
Protein co-detection
Combine whole transcriptome spatial analysis with immunofluorescence protein detection.
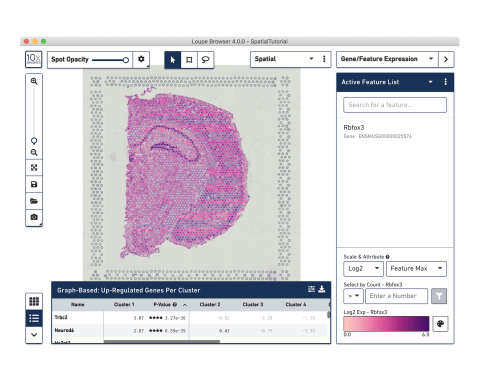
Streamlined data analysis
Combine histological and gene expression data with easy-to-use software.
Explore what you can do
Gain a complete view of disease complexity
Discover new biomarkers
Map the spatial organization of cell atlases
Identify spatiotemporal gene expression patterns
More than a tissue slide

FFPE
FFPE compatibility
Combine the benefits of histological techniques with the massive throughput and discovery power of RNA sequencing to detect the whole transcriptome in FFPE samples.
- Detect the whole transcriptome across entire FFPE tissue sections with high sensitivity and specificity
- Profile archival FFPE tissue samples to discover new biomarkers
- Compatible with two Capture Area sizes for more flexibility
Immunofluorescence
Protein co-detection
Enable discovery of novel cell types and their spatial organization by combining immunofluorescence and whole transcriptome spatial gene expression.
- Explore transcriptome and protein relationships with simultaneous spatial detection
- Discover new tissue biomarkers with cell-type specificity

Expanded sample access
Visium CytAssist-enabled workflow
With Visium CytAssist you can profile pre-sectioned tissues using the Visium for FFPE gene expression workflow.
- Start from FFPE blocks or pre-sectioned tissues on glass slides
- Pre-screen to find the most biologically significant tissue sections
- Integrates seamlessly with standard histology workflows
Proven results
Publications
Tertiary lymphoid structures generate and propagate anti-tumor antibody-producing plasma cells in renal cell cancer
Tertiary lymphoid structures generate and propagate anti-tumor antibody-producing plasma cells in renal cell cancer
Meylan M, et al. Immunity (2022).
Meylan M, et al. Immunity (2022).
Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro
Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro
Garcia-Alonso L, et al. Nat Genet (2021).
Garcia-Alonso L, et al. Nat Genet (2021).
Spatial transcriptomics of dorsal root ganglia identifies molecular signatures of human nociceptors
Spatial transcriptomics of dorsal root ganglia identifies molecular signatures of human nociceptors
Tavares-Ferreira D, et al. Sci Transl Med (2022).
Tavares-Ferreira D, et al. Sci Transl Med (2022).
Resources
Find the latest app notes and other Spatial Gene Expression documentation
Grant writing resource for Visium Spatial Gene Expression
Grant writing resource for Visium Spatial Gene Expression
Grant Application Resource, 10x Genomics
Grant Application Resource, 10x Genomics
Enriching pathological analysis of FFPE tumor samples with spatial transcriptomics
Enriching pathological analysis of FFPE tumor samples with spatial transcriptomics
App Note, 10x Genomics
App Note, 10x Genomics
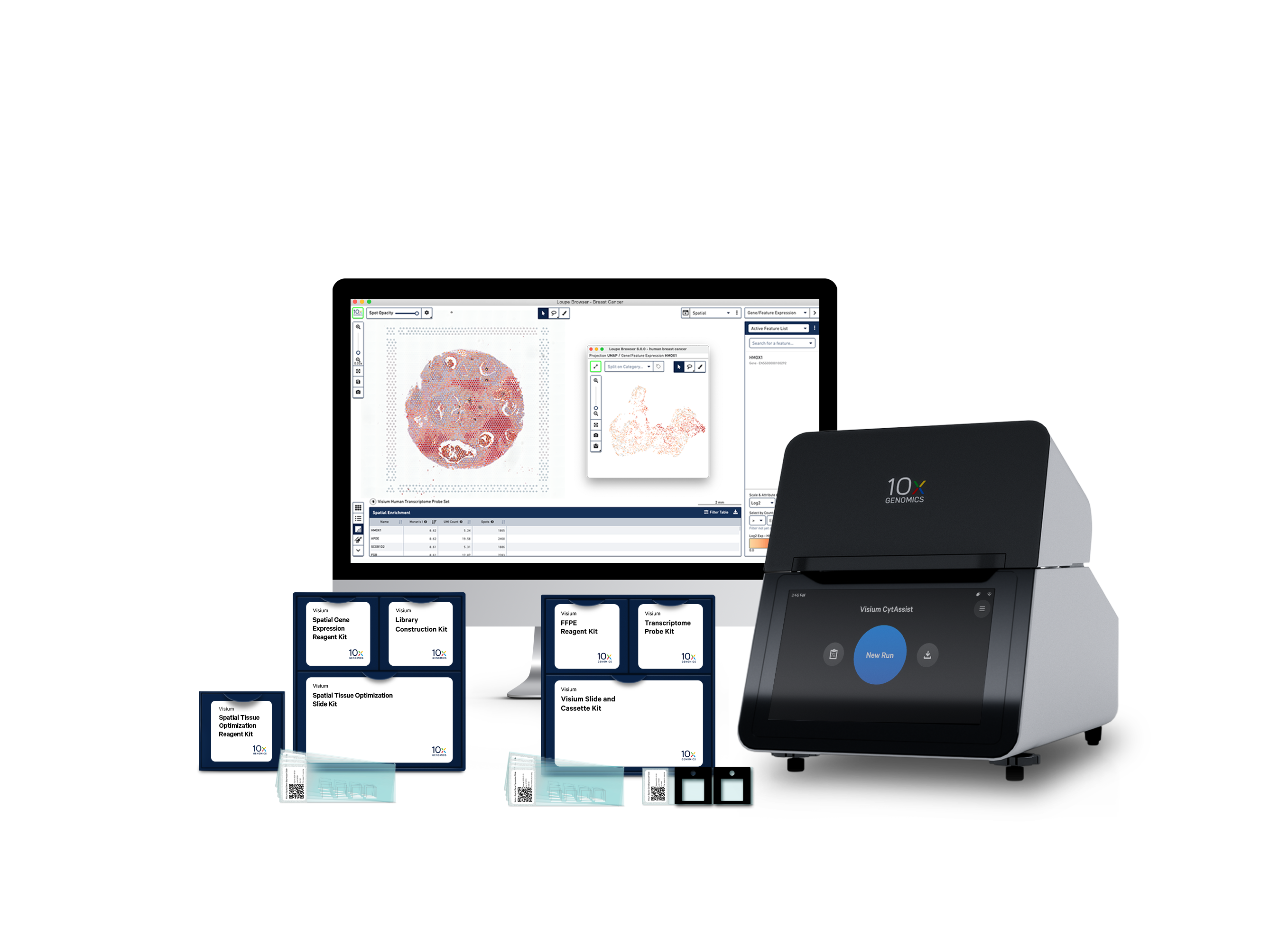
Our end-to-end solution

Visium Spatial capture slides
Easily adoptable within existing lab infrastructure, no need for a new instrument.

Visium Spatial Gene Expression reagents
Our reagent kits let you capture whole transcriptome gene expression with spatial resolution across an entire tissue section.
Analysis and visualization software
Our analysis pipelines
Our visualization software

World-class technical and customer support
Get the help you need with our expert support team, reachable by phone or e-mail.
Workflow
- 1
Prepare your sample
Embed, section, and place fresh frozen or FFPE tissue onto a Capture Area of the gene expression slide. Each Capture Area has thousands of barcoded spots containing millions of capture oligonucleotides with spatial barcodes unique to that spot.
Resources - 2
Stain and image the tissue
Use standard fixation and staining techniques, including hematoxylin and eosin (H&E) staining, to visualize tissue sections on slides using a brightfield microscope and immunofluorescence (IF) staining to visualize protein detection in tissue sections on slides using a fluorescent microscope.
Resources - 3
Permeabilize tissue and construct library
For fresh frozen tissue, the tissue is permeabilized to release mRNA from the cells, which binds to the spatially barcoded oligonucleotides present on the spots. A reverse transcription reaction produces cDNA from the captured mRNA. The barcoded cDNA is then pooled for downstream processing to generate a sequencing-ready library. For FFPE tissues, tissue is permeabilized to release ligated probe pairs from the cells, which bind to the spatially barcoded oligonucleotides present on the spots. Spatial barcodes are added via an extension reaction. The barcoded molecules are then pooled for downstream processing to generate a sequencing-ready library.
Resources - 4
Sequence
The resulting 10x barcoded library is compatible with standard NGS short-read sequencing on Illumina sequencers for massive transcriptional profiling of entire tissue sections.
Resources - 5
Analyze and visualize your data
Use our Space Ranger analysis software to process your spatial gene and protein expression data and interactively explore the results with our Loupe Browser visualization software.
Do I need to be a bioinformatician to use Loupe?
Loupe is a point-and-click software that’s easy for anyone to download and use.
Resources
Frequently asked questions
It allows you to perform spatial transcriptomics, which combines histological information with whole transcriptome gene expression profiles (fresh frozen or FFPE) to provide you with spatially resolved gene expression.
Visium is compatible with fresh frozen and FFPE tissues containing mRNA. We have separate kits that utilize different chemistries for fresh frozen and FFPE tissues. A list of tissues successfully tested in-house with the Visium assay can be found below.
Successfully tested tissues: Fresh frozen tissues | FFPE tissues
Visium is based on the Spatial Transcriptomics technology which was first reported in 2016. Since then Visium data has been published in nearly 90 peer-reviewed articles, including journals from Nature, Cell, and Science. Internally, over 30 different fresh frozen tissues and FFPE tissues have been tested successfully.
10x Genomics provides two types of software to help you analyze your data: Space Ranger and Loupe Browser. Space Ranger is an analysis software that automatically overlays spatial gene expression information on your tissue image and identifies clusters of spots with similar transcription profiles. You can then use Loupe Browser, a visualization software, to interactively explore the results. You can download Space Ranger and Loupe Browser from the 10x Genomics Support site at no cost.
The Visium Spatial Gene Expression for FFPE tissues assay does not require tissue optimization. However, the Visium Spatial Gene Expression assay for fresh frozen tissues does require tissue optimization due to differences in the chemistry and molecular biology between the Visium assays for the two sample preparation types.
Visium Spatial Gene Expression for fresh frozen and FFPE is compatible with hematoxylin and eosin H&E staining for morphological information and with IF staining for protein co-detection.